Hansen Solubility Parameters in Practice (HSPiP) e-Book Contents
(How to buy HSPiP)
Chapter 33, Into the 4th Dimension.
Donor/Acceptor
We
would like to warmly thank Professor Michael Abraham, University College
London, for his generous assistance with respect to his Abraham parameters.
As we’ve noted in other chapters, there are
other approaches to determining solubility. Each has its strengths and
limitations. Here is our view, in alphabetical order, of some of the main
approaches.
Abraham
parameters. For more than 20 years, Professor
Abraham and his team have methodically worked out a set of 5 parameters that
allow users to calculate solubilities and partition coefficients based on
linear free energy relationships. The parameters have been worked out through
careful, complementary experimental processes using solvatochromic shifts, NMR
shifts, GC and HPLC retention times. The approach has been adopted in a number
of areas and the large experimental database of parameters is a key aspect of
the approach. We will discuss the parameters later in this chapter.
COSMO-RS. Dr Andreas Klamt has developed an entirely new way of working with
solubility and partition issues. At the heart of the technique is quantum
mechanical calculations of each molecule (and, sometimes, each major conformer
of the molecule). Once the calculated data for the molecule is known,
subsequent calculations of interactions with other molecules are rapid. In
principle, therefore, COSMO-RS can do everything that all methods try to do,
but can do it from first principles. There is no doubt that this approach is
very powerful. As the base of validated quantum calculations increases the
usability for everyday problems will increase. For the purposes of this chapter
we note that COSMO-RS is able to generate “COSMOments” and a 5-parameter set
which we will discuss below.
MOSCED. MOSCED’s roots are not too distant from HSP. At an early stage it
was recognised that increasing the number and complexity of parameters would
provide better fits to the data. In particular, MOSCED introduced an acid/base
split of parameters to recognise specific interactions such as
acetone/chloroform. MOSCED becomes, therefore, a 5-parameter set. Recent MOSCED
papers have shown excellent agreement with activity coefficient data. However
there does not seem to be a robust set of MOSCED parameters for general use and
it is unclear to us how the current MOSCED parameters relate to the cohesive
energy density that is at the root of the concept.
UNIFAC and its variants. If you are a UNIFAC user then you probably aren’t
reading this eBook. UNIFAC’s strengths are unparalleled in its main domain of
use for vapour liquid equilibria in the chemical industry. However it doesn’t
seem to have caught on as a tool for the broad range of applications for which
HSP are so suitable. There is now a huge database of UNIFAC group coefficients
and if you have access to the database you can do many things better than you
can with HSP.
If we leave out UNIFAC as a special case,
it’s clear that other methods have 5 parameters whilst HSP has only 4. Why 4
– surely HSP has only 3 parameters? In this chapter we’ll emphasise the
fact that HSP calculations regularly use the MVol. MVol has appeared in all HSP
data tables and has been quietly working away in the background. The other 5
parameter sets also include a term that is equivalent to MVol.
An important paper, Andreas M. Zissimos, Michael H. Abraham, Andreas Klamt, Frank Eckert,
and John Wood, A Comparison between the Two General Sets of Linear Free
Energy Descriptors of Abraham and Klamt, J.
Chem. Inf. Comput. Sci. 2002, 42, 1320-1331, shows that the Abraham
approach and a simplified version of COSMO-RS (COSMOments) can both be
described by a 5-parameter set which can be adequately mapped between the two
techniques. This paper, incidentally, is an excellent introduction to both
approaches and is recommended for those who want to understand more about them.
For the purposes of this chapter, the paper
can be summarized by saying that the HSP terms of δD, δP and
MVol map onto corresponding terms in both Abraham and COSMOments. But δH is a
single term whilst Abraham has Acid/Base and COSMOments has Hdonor
and Hacceptor. The mapping of Abraham Acid/Base onto COSMOments Hdonor/Hacceptor
isn’t perfect – after all the other parameters aren’t perfect maps
– but the paper makes the point that the general idea of a 5-parameter
linear free energy space is a core thermodynamic concept.
Because it is clear that both Abraham and
COSMOments work well with extensive experimental databases it means that HSP
has to defend its use of a single δH parameter.
There has been no shortage of suggestions
that HSP should divide the δH parameter and there have been a few attempts
to make it happen. But there have been three practical objections. The first is
that the 4-parameter HSP (remember, we are including MVol as a parameter in
this chapter) works remarkably well. The second is that plotting in 4D space (δD, δP, δHD, δHA)
isn’t possible in this 3D world. The third is that there’s been no obvious way
to partion δH into the two terms.
But the classic acetone/chloroform case
where there is unambiguous evidence of donor/acceptor interactions between the
solvents shows that 4-parameter HSP cannot describe everything. However, as
noted in Charles’ history in the next chapter, even this case doesn’t show up
as special in general HSP use.
Acid/Base
or Donor/Acceptor
Sooner or later we have to decide on
terminology. The world is split into those who think that the best term to
describe the two terms is Acid/Base and those who think it should be
Donor/Acceptor. As you can see above, Abraham and Klamt use respectively
Acid/Base and Donor/Acceptor.
We decided that Acid/Base is rather too
literal so have chosen Donor/Acceptor. Hence we will talk of δHD and δHA
rather than δHA and δHB. The fact that the A of Acceptor is the opposite meaning to the A
of Acid is unfortunate, but there’s nothing we can do about it.
While we are clarifying terminology, let’s
restate that we talk about a 4-parameter or 5-parameter set but a 3D or 4D
viewing space. This is because the (scalar) MVol is not shown in the (vector)
plots of 3D or 4D space.
Inspired
by Abraham
Reading the extensive publications of the
Abraham team and examining their large public database of parameters it is
clear that their thoughtful approach to working out the Acid/Base parameters is
much to be admired. The IGC and HPLC techniques for HSP were developed
independently of the equivalent Abraham GC and HPLC work and show that in
principle the Abraham parameters could map onto an HSP 5-parameter set or, to
put it another way, HSP could in principle (though this won’t happen in
practice) gather Donor/Acceptor values in a similar fashion.
We therefore decided to create a
5-parameter HSP set using Abraham parameters to help us in one important step.
We decided to split δH using two rules.
Rule 1: δH²
= δHD² + δHA²
This rule ensures that everything about HSP
stays constant and we can always bring Donor/Acceptor back into 4-parameter
space without upsetting 40+ years of work.
Rule 2: For compounds with known Abraham
Parameters, δHD:δHA = Abraham Acid:Base
This rule allows us to get started on the
whole process of rationally splitting δH. We have no pure scientific justification for
this mapping other than our feeling that the Abraham approach to Acid/Base
determination (e.g. GC/HPLC) fits very well with HSP.
Once we were up and running with a basic
set of Donor/Acceptor splits, it was then possible to create Y-MB methods for
splitting molecules for which we had no Abraham parameters.
After that it requires a lot of checking
and invoking chemists’ common sense. If the automated process produced an amine
with a large δHD then clearly there was a problem with the process.
Out of this work came an important third
rule:
Rule 3: If you have no other way to decide,
make δHD=0 and δHA=δH.
This surprising rule is less surprising if
you glance at any Abraham table. For example, in a table of 500 compounds there
are 466 with Base values and 249 with a Acid values. And of those 249, only 96
have a value bigger than the Base value. So most molecules are Acceptors rather
than Donors. Of course Rule 3 should be used with care but note that it says
“if you have no other way to decide”. If a molecule has a carboxylic acid group
then you already know something so Rule 3 doesn’t apply.
Calculating
the distance
The previous section describes in a few
words a very large amount of work. This section describes some hard thinking.
When 5-parameter HSP seemed only an
impossible dream we assumed that if we had them it would be easy to calculate
the HSP distance. MOSCED, for example, uses (in our nomenclature) a term (δHD1-δHD2)(δHA1-δHA2)
instead of (δH1-δH2)². This term captures the
possibility of a “negative distance” which is what Donor/Acceptor is supposed
to accomplish.
However, we quickly found that we cannot
use such a term. If the two donor terms are equal then the first term is zero
so the distance is zero. But this cannot be the case for HSP. For example if δHD1
and δHD2 are both zero then the second term should be equal to the
classic δH distance because each δHA term equals the classic δH.
We
eventually found a distance formula that works well. It gives the sorts of
values we intuitively expect in all the test cases we can find. We would love
to be able to tell you that we understand the reason for the formula, but we
admit that we use it because (a) it seems intuitively right and (b) it is the
only formula out of many variants that gave us only values that made intuitive
sense. If someone from the HSPiP user community can prove it or provide a
better alternative we would be happy to acknowledge their work in a future
edition.
So if we
have (δHD1 , δHA1) and (δHD2 , δHA2)
then using the nomenclature Min(X,Y) to mean the minimum of X or Y, we define:
Equ. 1‑1 MinX1 = Min(δHD1 , δHA2)
Equ. 1‑2 MinX2 = Min(δHD2 , δHA1)
Equ. 1‑3 X1 = δHD1 - δHA2
Equ. 1‑4 X2 = δHD2 – δHA1
Equ. 1‑5 S1 = δHD1 - δHD2
Equ. 1‑6 S2 = δHA1 - δHA2
Equ. 1‑7 DA1 = Min(δHD1, δHA1)
Equ. 1‑8 DA2 = Min(δHD2 - δHA2)
Then
Equ. 1‑9 Distance = -Sqrt(MinX12+MinX22) + Min(Sqrt(X12+X22),
Sqrt(S12+S22))+Sqrt(DA1²+DA2²)
If both
δHD values =0 or both δHA values=0 then this term becomes the classic (δH1-δH2)²
If δHD1
is large and δHA2 is large while δHD2 is small and δHA1
is large then we have a classic donor/acceptor (chlororform/acetone) pair and
the distance is negative, which is precisely what we require. Mathematicians will note that when we
calculate the Distance² (in
the general distance formula) Distance² =Sign(Distance)* Distance².
Most
other cases show some reduction in distance from the classic δH distance
because of some favourable donor/acceptor interaction.
It’s
worth noting that although there are a large number of molecules with a high
δHA and a low (or zero) δHD there are very
few molecules (<5%) with a δHD more than twice δHA, and only 2% with more
than 3* δHA. So the likelihood of strong pure Donor:Acceptor effects is very
low.
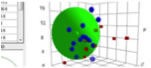
Our first 4D Spheres
We were
very nervous when we created our first Spheres using standard .hsd files (and
therefore known classic Spheres). What would happen if the results were very
different. Would this mean that 40 years of HSP have been wrong?
We
needn’t have worried. It quickly became clear that for most systems the results
were not very different. There are three very important reasons for this. The
first is obvious: because δD and δP have not changed, it’s not possible for the
Sphere to move too far. The second reason follows from the facts behind Rule 3
above. Because a large majority of molecules have a very low fraction of Donor,
the majority of interactions are Acceptor:Acceptor, in other words they are
simply classic δH. The third is simple statistics: the number of systems with
large δH values isn’t large and the effects of a Donor/Acceptor split from a
small δH value is not highly significant.
But the
fact that they are not very different
doesn’t mean that the whole 5-parameter approach is a waste of time. In some
cases the fits improve because there really are specific Donor/Acceptor
interactions for a few solvents and a few polymers, particles etc.
For us,
and we think for you, the move into 5-parameter space is a useful option. For
most work, the proven 4-parameter methodology works very well and there is no
need to add the complexity of the 5-parameter space. But given that we can
create the Donor/Acceptor split automatically and that the Sphere fitting isn’t
too much slower, it’s easy to explore the option if there are reasons to expect
that Donor/Acceptor effects will be large. And remember that “more” isn’t
always “better”. The split adds extra uncertainties to parameters which
themselves have errors. It is possible to make things worse by trying to be too
clever. So we recommend that for routine use, the 4-parameter classic method
should be your default. But feel free to enter the 4th dimension at
any time.
Reporting the results
Because
we cannot plot a 4D Sphere and because of Rule 3 above, we will continue to
plot the classic Sphere. So δH will always be calculated for you from δHD and
δHA via Rule 1 and plotted as normal. But of course we show you the δHD and δHA
values in a table so you can understand what is going on with each of the
solvents.
We
cannot save 4D data in the old .ssd file format. The new, standard, more
versatile .hsd format is equally happy with 3D and 4D data.
The problem of mixtures
One of
the great strengths of HSP is the ability to create arbitrary mixtures and
calculate the HSP by the standard mixing rule. Things get more complicated when
solvents are blended with Donor/Acceptor.
As an
extreme example (which, see the comments above, is highly unlikely to exist),
suppose we have a 50:50 blend of [10,0] and [0,10] – the mixture of an
equally strong pure donor and pure acceptor. In δH terms the classic answer is
5+5=10. One possible intuition is that the donor/acceptor cancel out giving
[0,0]. But it seems unlikely that the resulting blend would have no δH. It also
seems unlikely that it would be [10,10] with a resulting δH of Sqrt(10²+10²)=14.14.
After much experimentation we found that the simplest possible mixing rule, the
weighted average of δHD and δHA terms, gave sensible and intuitive results. In
this example the answer is [5,5] with a total δH of 7.07. Most other cases
aren’t so extreme and the resulting δH is not much reduced from the simple
weighted average of δH terms.
An
alternative possible rule is that δH of the mix remains the same as the classic
mixing rule, but the ratio of δHD and δHA is changed depending on the relative
values of the components. This is a slightly more complex rule to implement and
in most cases makes little difference, so for the moment we have chosen the
simple rule.
Because
so much of this is provisional, we’ve decided not to include donor/acceptor into the Solvent Optimizer.
Activity coefficients
In other
chapters we have pointed out that activity coefficients should be directly
calculable from HSP distance. But there is lots of experimental evidence to
show that this is not a reliable method for prediction. Clearly the
impossibility of a negative distance, indicating a Donor/Acceptor interaction,
has been a limitation of the 4-parameter HSP.
For this
release of HSPiP we simply haven’t had the time to do the vast amount of work
to see if a 5-parameter model gives more reliable predictions. That is
something for the next release.
Use with care
We
believe that this shift to a 5-parameter HSP is a necessary step in the
evolution of HSP. But our evidence is that the step is in most cases not of great importance. So classic HSP has not
suddenly been invalidated. Therefore we are not shifting everything at once. On
the surface, much remains 4-parameter, though behind the scenes we use
5-parameter when it seems sensible. Unless you select the D/A option you won’t
even see the 5-parameter data.
We have
some internal cases where the Donor:Acceptor correction is so large, and the δD and δP
parameters are so close that the distance is negative, though without an
especially high solubility. In other words our formula seems to over-correct in
some cases. We will carry on our internal arguments on the issue while this
current edition is out in the open. We very much look forward to your input to
the debate.
But the
key take-home message here is to “use with care”. It’s possible that we will
change the Distance algorithm and the Mixing algorithm. Our splitting ratios
may change with time as we gather more experimental data. So enjoy exploring
the 4th dimension, take what value you can from it, but, at this
stage, don’t rely too heavily on its predictions.
E-Book contents | HSP User's Forum