HSP Application note #46
Green Solvent and Hansen Solubility Parameter (HSP)2010.9.24
HSPiP Team Senior Developer, Dr. Hiroshi Yamamoto
Green Solvent=Environmentally friendly solvents
Non-Flammable
VOC regulation(Volatile Organic Compounds regulation)
HAPs regulation(Hazardous Air Pollutions regulations) US
Kashin-Hou(Japanese regulation for new chemicals)JP
Reach ( registration, evaluation and authorization of chemicals) EU
TSCA (Toxic Substance Control Act) US
Toxicity (LD50 etc.)
ODP (Ozone Depression Potential, Montreal protocol)
GWP (Global warming Potential, Kyoto protocol )
Regulation free solvent (design of solvents mixture)
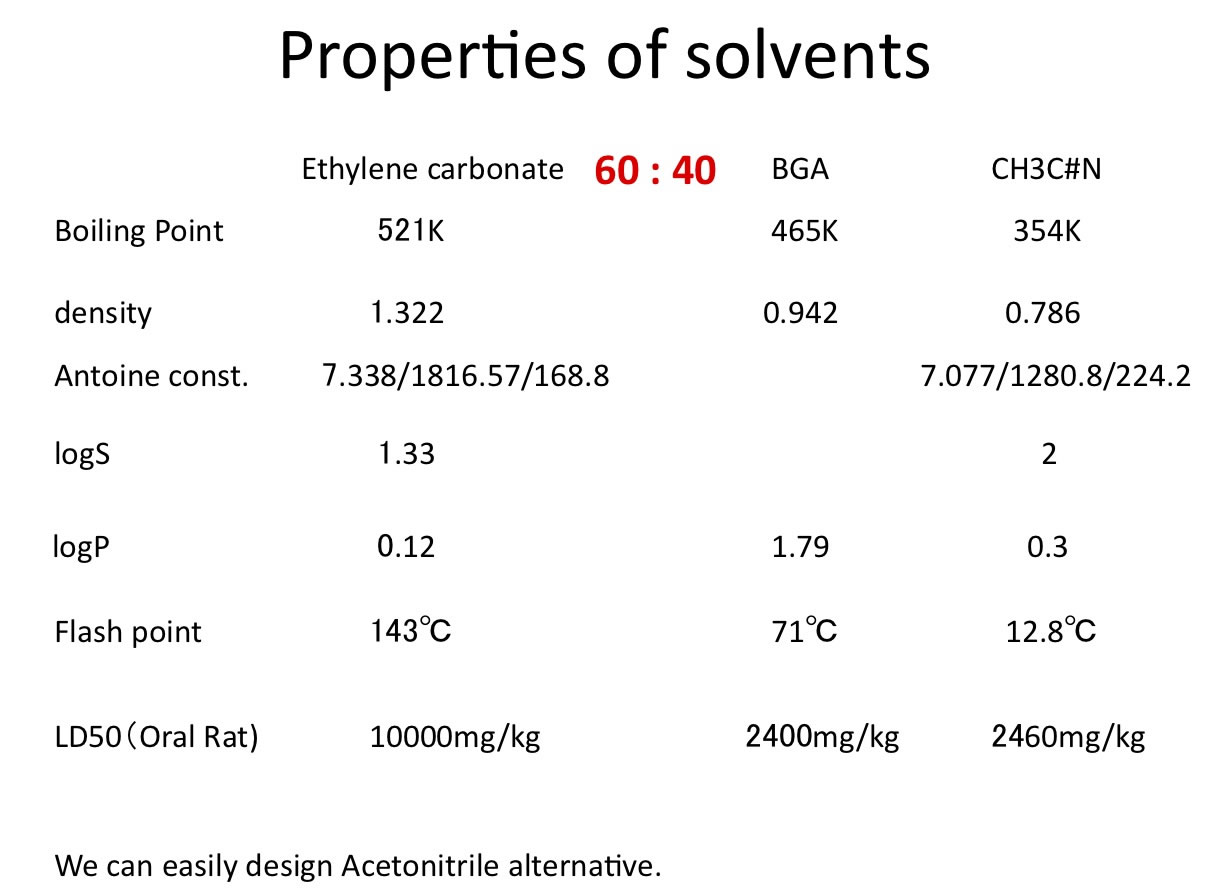
Ex. Search Acetonitrile alternatives
In organic synthesis, Acetonitrile solvent is very popular.
How can we search Acetonitrile alternatives using HSP?
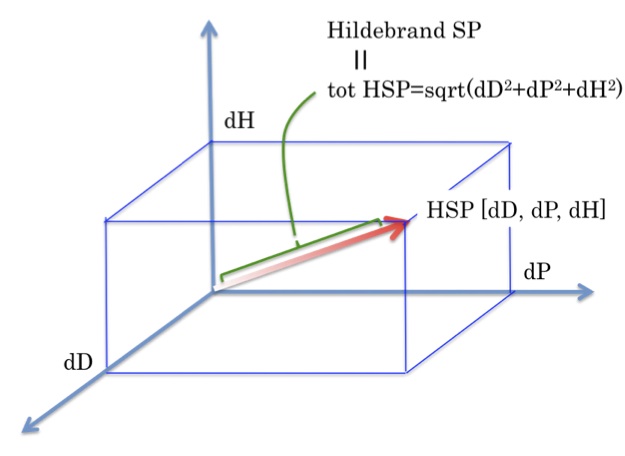
Hansen Solubility Parameters (HSP)Hansen Solubility Parameters(HSP) were developed by Charles M. Hansen as a way of predicting if one material will dissolve in another and form a solution. They are based on the idea that "like dissolves like" where one molecule is defined as being 'like' another if it bonds to itself in a similar way.
What can perhaps be surprising is that one can assign HSP to so many different things. Gases like carbon dioxide, solids like carbon-60, sugar, and biological materials like human skin, depot fat, DNA, and even some proteins all have HSP. The list can be continued with drugs, polymers, plasticizers, and in fact any organic material and even many inorganic materials like salts. The only requirement for an experimental confirmation is that the material must behave differently in a sufficient number of test solvents upon contact. Pirika JAVA Demo Applet calculate HSP. HSPLight is available here. |
1. Acetonitrile HSP[15.3, 18, 6.1] dP is very large. dD is small.
2. Search from most common 81 solvents
| 10 Acetonitrile | 15.3 | 18 | 6.1 | Ra |
| 115 gamma-Butyrolactone (GBL) | 18 | 16.6 | 7.4 | 5.73 |
| 535 1-Nitropropane | 16.6 | 12.3 | 5.5 | 6.29 |
| 608 Sulfolane (Tetramethylene Sulfone) | 18 | 18 | 9.9 | 6.60 |
| 363 Ethylene Carbonate | 18 | 21.7 | 5.1 | 6.62 |
| 303 Dimethyl Sulfoxide (DMSO) | 18.4 | 16.4 | 10.2 | 7.60 |
| 7 Acetone | 15.5 | 10.4 | 7 | 7.66 |
gamma-Butyrolactone is the nearest, even though HSP distance(Ra) is 5.73. Not so good.
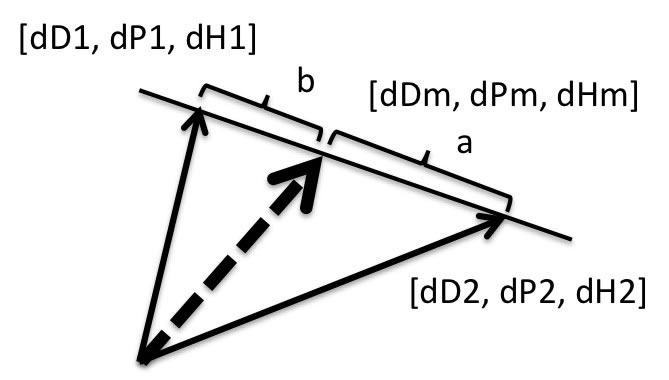
3. Search Solvents mixture
From the combination of 81*80 and change ratio.
| A | A ratio | B | B ratio | dDm | dPm | dHm | Ra |
| 7:Acetone | 0.5 | 363:Ethylene Carbonate | 0.5 | 16.75 | 16.05 | 6.05 | 3.49 |
| 363:Ethylene Carbonate | 0.7 | 963:t-Butyl Acetate | 0.3 | 17.1 | 17.05 | 5.43 | 3.78 |
| 363:Ethylene Carbonate | 0.6 | 369:Butyl Glycol Acetate | 0.4 | 16.92 | 16.02 | 5.78 | 3.81 |
| 363:Ethylene Carbonate | 0.7 | 666:Texanol | 0.3 | 17.13 | 17.02 | 6.51 | 3.81 |
| 363:Ethylene Carbonate | 0.7 | 440:Iso-Propyl Acetate | 0.3 | 17.07 | 16.54 | 6.03 | 3.83 |
Searched Ethylene carbonate base solvents mixture.
HSP of Solvents Mixture[dDm, dPm, dHm]=[(a*dD1+b*dD2), (a*dP1+b*dP2),(a*dH1+b*dH2)]/(a+b)
Volume base ratio. Pirika Java demo applet design solvents mixture. GSD is available here. |
Which one is the first priority?
Classification of hazardous compounds in Japan
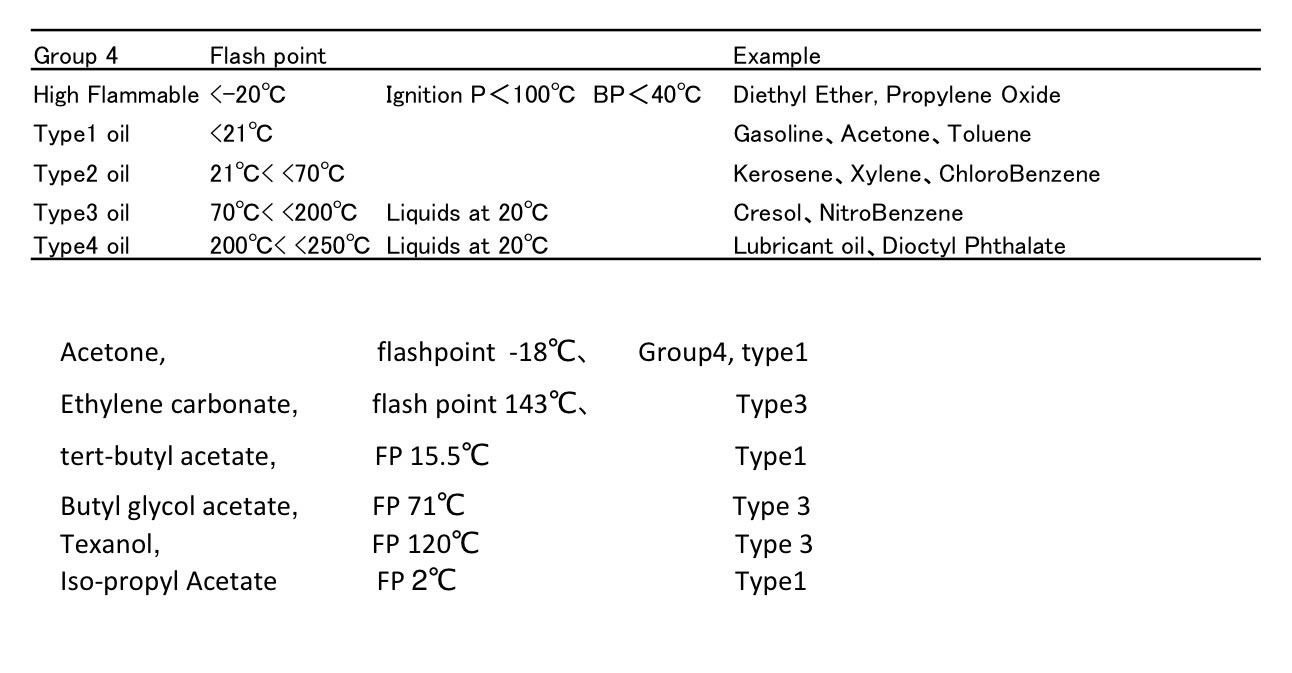
Flash point is very important property.
HSPiP(Hansen Solubility Parameters in Practice)The first edition of HSPiP that appeared in November, 2008, greatly enhanced the usefulness of the Hansen solubility parameters (HSP). The HSP values of over 1200++ chemicals and 500 polymers are provided in convenient electronic format and have been revised and updated using the latest data sources in the second edition (March, 2009). A third edition of the HSPiP appeared in March, 2010. There are now 10,000 compounds in the HSP file which also includes data on density, melting point, boiling point, critical parameters, Antoine constants and much more. The user is able to carry out many different sorts of optimisations of solubility, evaporation, diffusion, adhesion, create their own datasets (automatically if required) and explore the huge range of applications for HSP in coatings, paints, nanoparticles, cosmetics, pharma, organic photovoltaics and much more. The 3rd Edition v3.1 was released on 12 December 2010. Current users can upgrade free (now v3.1.09) by downloading the latest .msi installer from http://hansen-solubility.com The 4th Edition v4.0.x was released on 2 Jan. 2013. The Current users can upgrade with free charge. 2013.1.28 The Visual How to manual of HSPiP. You can understand what HSPiP can do. |
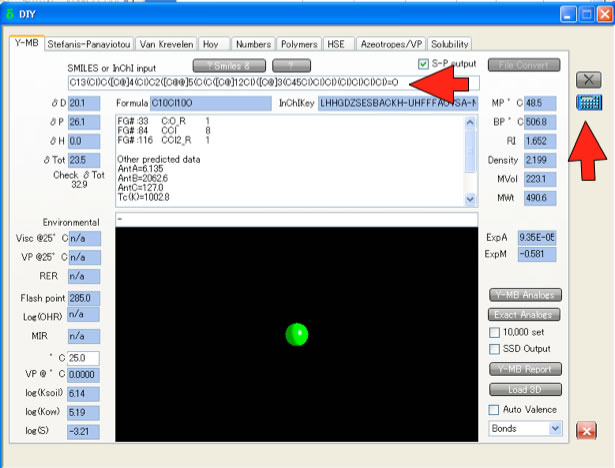
If you have Smiles structure and HSPiP software, Y-MB function will calculate HSP and other properties immediately.
Smiles(Simplified Molecular Input Line Entry Syntax) SMILES is a string obtained by printing the symbol nodes encountered in a depth-first tree traversal of a chemical graph. Pirika JAVA Demo Applet getting Smiles. Draw2Smiles is available here. |
Y-MB Properties EstimationY-MB break Smiles into correspponding Functional Groups and Estimate various Properties. These estimation schemes are come from Pirika technologies.
Pirika JAVA Demo Applet calculate Properties. PirikaLight is available here. |